microCLIP
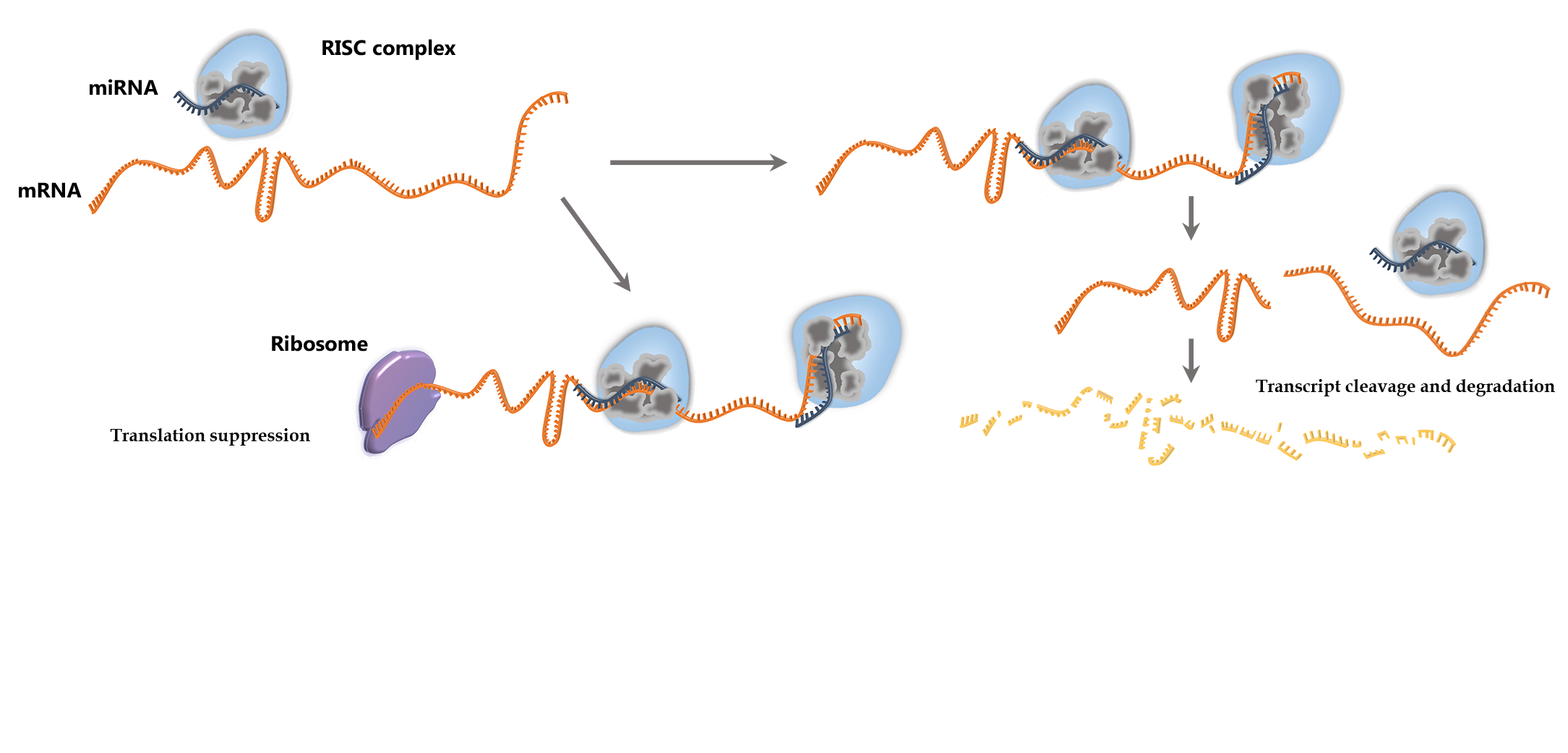
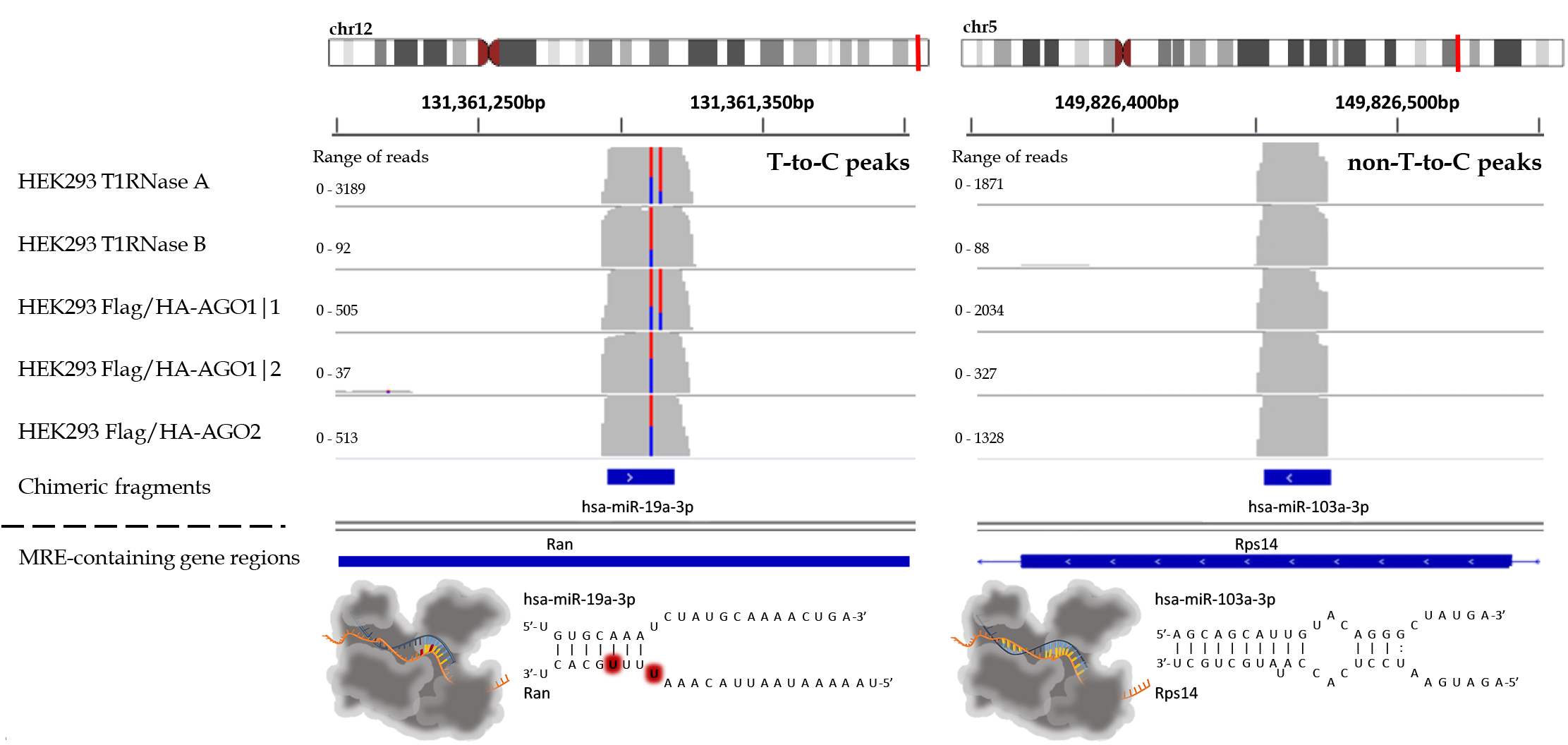
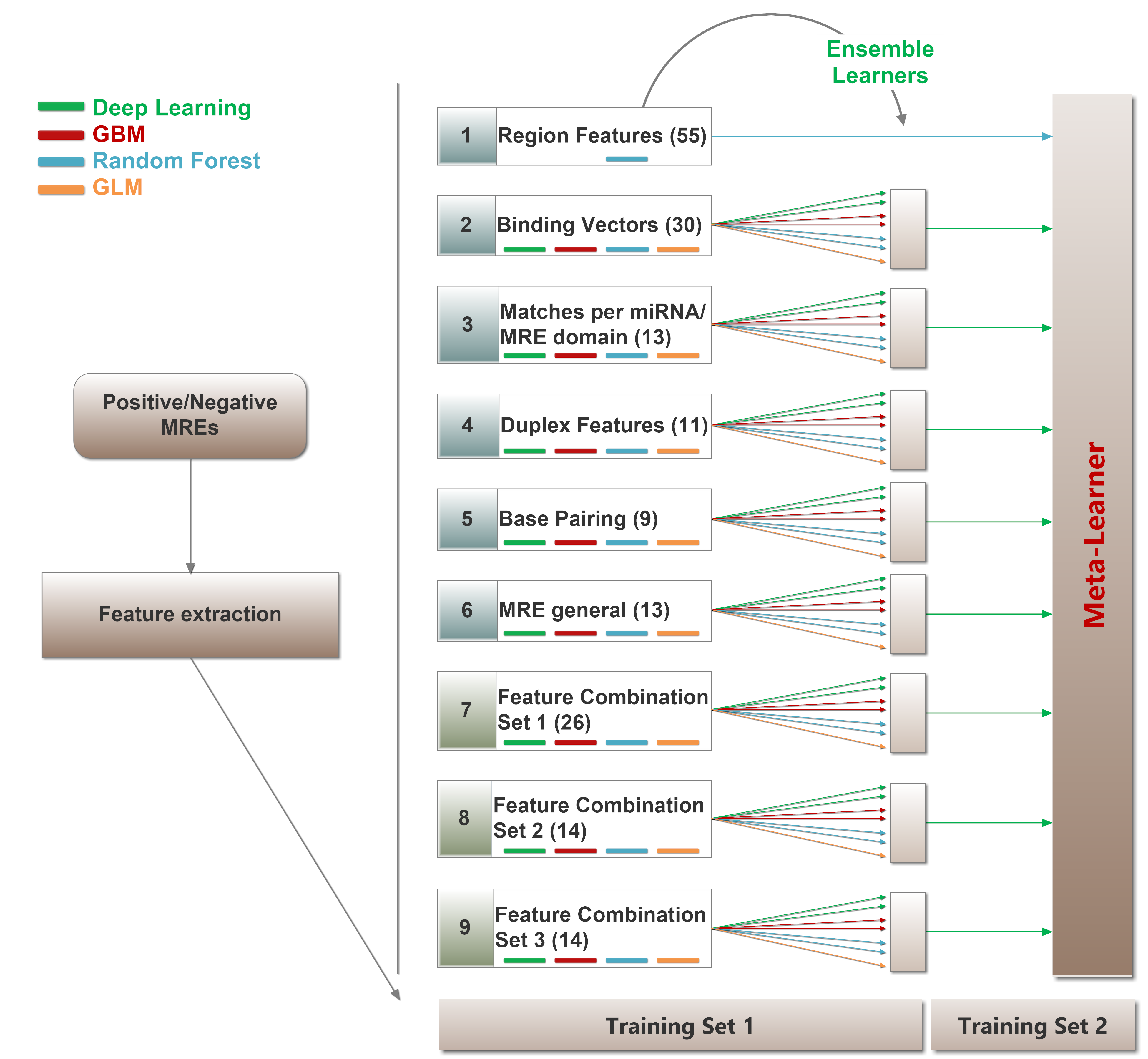
microCLIP algorithm can be used for the identification of transcriptome wide miRNA-target interactions. It operates on AGO-PAR-CLIP sequencing reads, requiring a SAM/BAM alignment file and a list of miRNAs as minimum input. microCLIP framework combines deep learning classifiers under a super learning scheme for CLIP-guided detection of miRNA interactions. It extracts features for each candidate miRNA Recognition Element (MRE) and respective AGO-bound enriched regions/clusters. It subsequently scores MRE sites through a multi-layer classification scheme. The output of the algorithm is a BED-like file comprising (non-)canonical miRNA interactions.
microCLIP is a standalone framework developed in R, while further details concerning the installation and sample files can be accessed upon clicking relevant links.
Please Cite:
Paraskevopoulou, M.D., Karagkouni, D., Vlachos, I.S. et al. microCLIP super learning framework uncovers functional transcriptome-wide miRNA interactions. Nat Commun 9, 3601 (2018). https://doi.org/10.1038/s41467-018-06046-y